Fitting for the photometric parameters of MUSE images
The photometric parameters of one or more MUSE exposures of a field
can be fit either using the fit_photometry script, or via one or
more provided python functions. The command-line options of the
fit_photometry script can be seen by passing it the -h argument:
% fit_photometry -h
usage: fit_photometry [-h] [--regions file-star-notstar-or-none]
[--star ra dec radius] [--segment] [--fix_scale factor]
[--fix_bg offset] [--fix_dx arcsec] [--fix_dy arcsec]
[--fix_fwhm arcsec] [--fix_beta value]
[--hst_fwhm arcsec] [--hst_beta value] [--margin arcsec]
[--save] [--taper pixels] [--extramask text] [--display]
[--nowait] [--hardcopy format-or-none] [--title text]
[--apply] [--resample] [--verbose]
hst_image muse_images [muse_images ...]
positional arguments:
hst_image The name of the FITS file containing the reference
HST image. This must have been decimated and
resampled onto the same coordinate grid as the MUSE
images. The HST bandpass filter used to obtain this
image should match the bandpass of the MUSE images as
closely as possible. Note that the `regrid_hst_to_muse`
script or the `mpdaf.obj.Image.align_with_image()`
function can be used create a subimage of an HST
image that has the same coordinate grid as a MUSE
image.
muse_images One or more FITS files containing MUSE images. These
should all be images of the same field and bandpass
as the HST image named by the --hst argument. Note
that the `make_wideband_image` script or the
`mpdaf.obj.Cube.bandpass_image()` can be used to
extract an image of a given HST bandpass from a MUSE
cube.
optional arguments:
-h, --help show this help message and exit
--regions file-star-notstar-or-none
DEFAULT=star
This can be "none", to indicate that all pixels
should be used in the global image fit, "star" to
restrict the fit to pixels within any optional circle
defined by the --star argument, "notstar" to restrict
the image fit to pixels outside any circle defined by
the --star argument, or the filename of a ds9 region
file.
This option can be used to exclude damaged regions of
an image, or to exclude sources that would degrade
the global PSF fit, such as saturated stars, stars
with significant proper motion, or variable sources.
Alternatively this option can also be used to
restrict the fit to one or more objects, by masking
everything except small regions around these objects.
Only ds9 circle, ellipse and box regions are
supported. Other types of ds9 regions and
configuration parameters, are simply ignored. For
each region, be careful to tell ds9 whether you want
the region to be included or excluded. Also be
careful to specify either fk5 or physical pixel
coordinates.
If the value of this argument is "star", and the
--star argument has also been provided, then a region
is substituted that only includes the circular region
specified by the --star argument.
Similarly, if "notstar" is passed, and the --star
argument has been provided, then a region is
substituted that excludes the circular region set by
the --star argument.
--star ra dec radius DEFAULT=None
Perform photometry fits to a star at a specified
position. This is done in addition to the global
image fitting procedure, so two sets of
photometry results are reported when this option
is selected.
The option expected 3 values. These are the right
ascension and declination of the star in decimal
degrees, and the radius of the area over which to
perform the fit, in arcseconds.
If the --regions argument is also present, and
its value is the word "star", then the imaging
fitting procedure is also restricted to the
region of this star. This can be used as a
consistency check, as both methods should yield
similar results.
--segment Ignore areas that don't contain significant objects
by ignoring pixels that are below the median value in
a morphologically opened version of the HST image.
--fix_scale factor DEFAULT=None
Use this option to fix the calibration scale
factor, (MUSE_flux / HST_flux) to the specified
value while fitting. The default value is "none",
which means that the parameter will be fitted.
--fix_bg offset DEFAULT=None
Use this option to fix the calibration zero-offset
(MUSE_flux - HST_flux) to the specified value while
fitting. The default value is "none", which means
that the parameter will be fitted.
--fix_dx arcsec DEFAULT=None (arcseconds)
Use this option to fix the x-axis pointing offset,
(MUSE_x - HST_x) to the specified value while
fitting. The default value is "none", which means
that the parameter will be fitted.
--fix_dy arcsec DEFAULT=None (arcseconds)
Use this option to fix the y-axis pointing offset,
(MUSE_y - HST_y) to the specified value while
fitting. The default value is "none", which means
that the parameter will be fitted.
--fix_fwhm arcsec DEFAULT=None (arcseconds)
Use this option to fix the FWHM of the Moffat PSF
to the specified value while fitting. The default
value is "none", which means that the parameter
will be fitted.
--fix_beta value DEFAULT=2.5
Use this option to fix the beta exponent of the
Moffat PSF to the specified value while fitting.
The default value is 2.5. Change this to "none"
if you wish this parameter to be fitted.
--hst_fwhm arcsec DEFAULT=0.085 (arcseconds)
The FWHM of a Moffat model of the effective PSF of
the HST. The default value that is used if this
parameter is not specified, came from Moffat fits to
stars within HST UDF images. To obtain the closest
estimate to the dithered instrumental PSF, these fits
were made to images with the smallest available pixel
size (30mas).
--hst_beta value DEFAULT=1.6
The beta parameter of a Moffat model of the effective
PSF of the HST. The default value that is used if
this parameter is not specified, came from Moffat
fits to stars within HST UDF images, as described
above for the hst_fwhm parameter. This term is
covariant with other terms in the star fits, so there
was significant scatter in the fitted values. From
this range, a value was selected that yielded the
least scatter in the fitted MUSE PSFs in many MUSE
images from different MUSE fields and at different
wavelengths.
--margin arcsec DEFAULT=2.0 (arcseconds)
The width of a margin of zeros to add around the
image before processing. A margin is needed because
most of the processing is performed using discrete
Fourier transforms, which are periodic in the width
of the image. Without a margin, features at one edge
of the image would spill over to the opposite edge of
the image when a position shift was applied, or when
features were widened by convolving them with a
larger PSF. The margin width should be the maximum of
the largest expected position error between the two
input images, and the largest expected PSF width.
--save Save the result images of each input image to FITS
files.
--taper pixels DEFAULT=9 (pixels)
This argument controls how transitions
between unmasked and masked regions are
softened. Because the fitting algorithm
replaces masked pixels with zeros, bright
sources that are truncted by masked regions
cause sharp changes in brightness that look
like real features and bias the fitted
position error. To reduce this effect,
pixels close to the boundary of a masked
region are tapered towards zero over a
distance specified by the --taper argument.
The method used to smooth the transition
requires that the --taper argument be an odd
number of pixels, so if an even-valued
integer is specified, this is quietly
rounded up to the next highest odd
number. Alternatively, the softening
algorithm can be disabled by specifying 0
(or any value below 2).
--extramask text DEFAULT=None
If the value of this argument is not the
word "none", then it should name a FITS file
that contains a mask image to be combined
with the mask of the MUSE image.
Specifically, this FITS file should have an
IMAGE extension called 'DATA' and the image
in that extension should have the same
dimensions and WCS coordinates as the MUSE
images. The pixels of the image should be
integers, with 0 used to denote unmasked
pixels, and 1 used to denote masked pixels.
--display Display the images, FFTs and star fits, if any.
--nowait Don't wait for the user to interact with each
displayed plot before continuing.
--hardcopy format-or-none
Write hardcopy plots of the fitting results to files
that have the specified graphics format (eg. "pdf",
"jpg", "png", "eps"). Plots of the fits will be
written to filenames that start with the name of the
MUSE input file, after removing any .fits suffix,
followed by either "_image_fit.<suffix>" for the plot
of the image fit, or "_star_fit.<suffix>" for plots
of any star fits.
--title text DEFAULT=None
Either a plot title, "none" to request the default
title, or "" to request that no title be displayed
above the plots.
--apply Derive corrections from the fitted position errors
and calibration errors, apply these to the MUSE
image, and write the resulting image to a FITS
file. The name of the output file is based on the
name of the input file, by replacing its ".fits"
extension with "_aligned.fits". If the input muse
image was not read from a file, then a file called
"muse_aligned.fits" is written in the current
directory. Also see the --resample option.
--resample By default the --apply option corrects position
errors by changing the coordinate reference pixel
(CRPIX1,CRPIX2) without changing any pixel values.
Alternatively, this option can be used to shift
the image by resampling its pixels, without
changing the coordinates of the pixels.
--verbose Report details of each fit, including chi-squared,
correlations, etc. Normally only summaries of the
fitted parameters are displayed.
%
General Usage
The fit_photometry script fits for the photometric parameters of one or more MUSE images of a field. It does this by convolving a specified HST image of the same field, with a PSF that best reproduces the resolution of the MUSE image; also by scaling and offsetting its fluxes with numbers that best reproduce the fluxes of the MUSE image, and finally also by shifting the position of the HST image by a vector amount that best lines up features in the two images. The fitting process is undertaken in the Fourier plane, where convolution and image shifts are simple multiplications. The goodness of the fit is likewise determined by comparing corresponding pixels of the FFTs of the MUSE image and the HST image.
By default, the script performs the above procedure on each of the
MUSE images in turn, and reports a summary of the fitted photometry
parameters. A more verbose report of the fit results can be requested
using the --verbose option. When there are multiple MUSE images to
be fit, the script automatically distributes the work between multiple
processes to make it more efficient on multi-processor computers. The
results are always reported in the order that the MUSE images are
given on the command-line, regardless of the order in which they are
actually completed in the different processes.
Plotting options
By adding the –display option, the script can be asked to plot the pre-processed MUSE and HST images and the best-fit images, along with their FFTs. By default, these plots wait for the user to interact with them and dismiss them, before the fitting process resumes. Alternatively, the –nowait option makes the plots non-interactive. In this case they disappear automatically as soon as the script ends, or as soon as a new plot is plotted, such as a star plot (see below), or the plots of a subsequent MUSE image.
Beware that the plots are generated and displayed by the background worker processes, as the fits are completed. This means that when multiple MUSE files are being processed, there may be many plots displayed simultaneously. It also means that the plots may be displayed in a different order to the textual results, which are reported in the order that the images are given on the command line. As such, it is better to restrict the use of the –display option to when only one MUSE image is being processed by the fit_photometry script. Alternatively, the plots can be saved as PDF files for later inspection, by using the –hardcopy option instead of the –display option.
Regions
By default the fit_photometry script performs a
global fit between an HST image and each MUSE image. However sometimes
it is better to restrict the fit to a bright star. At other times it
can be better to fit to everything except a bright object that is
confusing the fit, such as an object that has changed in position or
in flux since the HST image was taken. This is the purpose of the
--regions argument.
The --regions argument has three possible arguments:
none
This tells fit_photometry to perform the fit to all the pixels of the MUSE and HST images.
star
This is described in more detail under Star fitting. It tells the script to restrict the fit to a region of pixels defined by the
--starargument.
filename
This should be the name of a ds9 region file. Ds9 region files are generated by the ubiquitous ds9 FITS viewing program. The simplest way to generate a suitable file is to run ds9 on either a MUSE exposure, or the corresponding HST image.
The ds9 program can be used to interactively select regions of various shapes, and save the resulting shapes to a ds9 region file. Of the available shapes, only the circle, ellipse and box shapes are supported by fit_photometry. Other regions that are found in regions files by fit_photometry are simply ignored. The fit_photometry script also heeds the
fk5andphysicalcoordinate system designators. If no coordinate system designator is found before the first region definition, that region is assumed to be specified inphysicalcoordinates, which are FITS 1-relative pixel indexes. Other coordinate systems, such as B1950, are recognized, but they elicit an error message from fit_photometry and terminate the script.By default, ds9 creates include regions. For example, an circular include region indicates that fit_photometry should restrict its fitting process to the interior of the circular region. Alternatively, ds9 can be asked subsequently create exclude regions by pulling down the Region menu and selecting
Excludefrom thePropertiessub-menu. Note that ds9 distinguishes excluded regions from included regions by drawing a red diagonal line across them.In fit_photometry if all of the regions in a region file are include regions, then the fit will be restricted to the pixels within those regions.
Alternatively, if all of the regions are exclude regions, then the fit will be restricted to all of the image pixels except for those within the excluded regions.
If the region file contains both include and exclude regions, then the fit is performed on all pixels that are outside the excluded regions, plus any pixels that are within both the included and excluded regions.
Region files can also be generated by hand or automatically by a python script. For example, a file that is given the following contents:
fk5; circle(53.157969, -27.769193, 2.5")This would tell fit_photometry to only perform the imaging fit to pixels within 2.5 arcseconds of a point at an fk5 Right Ascension and declination of 53.157969, -27.769193 degrees respectively. This line is actually the contents of a region file that is used for selecting a small region around a bright star in the MUSE UDF01 field. Carefully note the
"after the 2.5 at the end of the line. This character indicates that the radius of the circle is 2.5 arcseconds. Without this character, the radius would be assumed to be in degrees.The above line restricts the fit to an area around a star. To alternatively exclude the same area, so that the fit is performed to all pixels except those of the star, the file could instead contain the following line:
fk5; -circle(53.157969, -27.769193, 2.5")Note the minus sign before the shape name. This is what indicates that a region should be excluded from the fit. For completeness, the contents of the following region file would exclude two bright stars from the fit:
fk5 -circle(53.157969, -27.769193, 2.0") -circle(53.162822, -27.767150, 2.0")
Region files for the MUSE UDF fields
The following region files are provided for use with the MUSE UDF fields.
Region file
Purpose
Exclude bright stars and QSOs
Select the bright star in UDF01
Select the bright QSO in UDF01
Select the bright star in UDF04
Select the bright star in UDF05
Select the bright star in UDF06
Select the bright star in UDF07
Star fitting
As described above under Regions, one way to restrict the
fitting process to a single star, is to use the --regions option
to specify a region that excludes all of the image from the fit, apart
from a small area around the star. However the
–star option provides a more convenient way to
do this that doesn’t require the creation of a region file. It also
selects a second form of photometry fitting, specific to stars.
The –star option takes 3 arguments which define a circular region around the position of a star. These are the Right Ascension and Declination of the center of the region, in degrees, and the radius of the region in arcseconds. By default this elicits two changes to the way that fit_photometry proceeds:
If the –regions parameter has the value
star(the default), then the image fit is limited to pixels inside the circular region specified to the –star.Alternatively, if the –regions parameter has the value
notstar, then the image fit is limited to pixels outside the circular region specified to the –star.Finally, if the –regions parameter has the value
noneor is given the name of a ds9 region file, then the image fitting procedure proceeds as though the –star argument had not been specified.In addition to the above changes, the –star option also elicits a second photometric fitting method, which provides an independent estimate of the photometric parameters.
This algorithm separately fits Moffat profiles to a given star in the MUSE image and the HST image. The final reported values for the PSF of the MUSE image are the fitted FWHM and beta values of the Moffat profile in the MUSE image. The reported value for the flux scale-factor is the ratio of the integrated Moffat fluxes fitted in the MUSE and HST images. Similarly, the reported flux zero-offset is the difference between the zero-offsets of the MUSE and HST Moffat fits, and the pointing offset of the MUSE image is the difference between the centroids of the fitted Moffat profiles in the two images.
The results of the normal image fitting algorithm and the star fitting algorithm are reported one after another. Similarly, when the –display and/or –hardcopy options are selected, separate plots are displayed for the image fitting technique and for the star-profile fitting technique.
The fitted parameters
The fitted parameters are named as follows:
Parameter
Description
fwhm
The PSF full-width at half-maximum
beta
The PSF Moffat beta value
scale
The flux scale (MUSE_flux / HST_flux)
bg
The flux offset (MUSE_flux - HST_flux)
dx
The X-axis offset (MUSE_x - HST_x)
dy
The Y-axis offset (MUSE_y - HST_y)
Note that the MUSE PSF is modeled as a 2D Moffat function. A 2D Moffat profile with a total integrated flux of 1, is defined as follows
In this equation, \(\beta\) is the beta parameter listed as one of the fitted parameters, and \(\alpha\) can be calculated from the FWHM, \(w\), as:
The following figure shows a comparison of a Moffat profile of \(\beta=2.5\) to a Gaussian profile of the same FWHM:
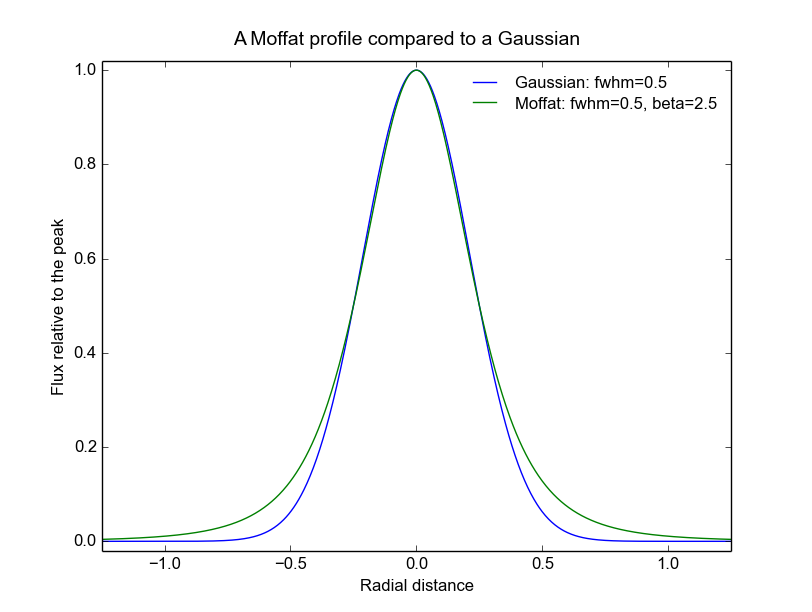
In general, near the peak of the PSF, Moffat profiles are similar to Gaussian profiles. However they have wider wings, as can be seen in the plot. As beta increases, the strength of the wings steadily decreases and the function becomes more and more like a Gaussian.
Indicating which parameters are to be fitted
For each photometric parameter, par, there is a corresponding option
called –fix_par. If this option is assigned a floating point
value, then the parameter par is held at that value during the
least-squares fitting process. For example, to force the Moffat beta
parameter to have a value of 2.8, one would use --fix_beta=2.8,
whereas to let it vary during the fit, one would use
--fix_beta=none.
By default, the fwhm, scale, bg, dx and dy parameters are fitted, but
the beta parameter is held at the value 2.5. This is done because
there is significant covariance between this parameter and other
parameters in the fit, and it is easier to obtain a smooth trend in
the fitted FWHM value versus wavelength if the beta value is
constrained to the same value in all of the fits. Star profile fits
indicate that common values of beta in MUSE images are between about
2.2 and 3.0, depending on the seeing conditions. The restriction on
beta can be removed, as described below, by adding the option
--fix_beta=none. Alternatively, it can be fixed to another value.
Writing a corrected MUSE image
If the optional --apply argument is included in the argument list to
fit_photometry, then the MUSE image is
corrected for the fitted pointing and calibration errors, and the
resulting image is written to a new FITS file, which has the name of
the MUSE input file, but with the “.fits” extension replaced by
“_aligned.fits”. By default, the fitted pointing errors are corrected
by changing the coordinate reference pixel (CRPIX1, and CRPIX2) in the
FITS header. Alternatively, the pointing can be corrected by
resampling the image, to shift it without changing the coordinates of
the pixels. This option is requested by including --resample in the
argument list, in addition to --apply.